
Integrated Predictive Toxicology & QSAR Modeling Platform
OrbiTox is a predictive toxicology platform that combines extensive, expertly curated data with hundreds of validated QSAR models. The platform integrates over 850K chemicals and over 500K high-throughput screening endpoints with interconnected information from chemical, genetic, and biological domains. Advanced read-across analysis and state-of-the-art machine learning, alongside interpretable descriptors, fill data gaps and deliver actionable predictions for mechanistic insights and chemical design. Additionally, OrbiTox offers an intuitive, secure interface that lets users efficiently analyze both public and proprietary data.
Interpretable QSAR Models Backed by an Extensive Curated Multi-Domain Database
Interpretable QSAR Models
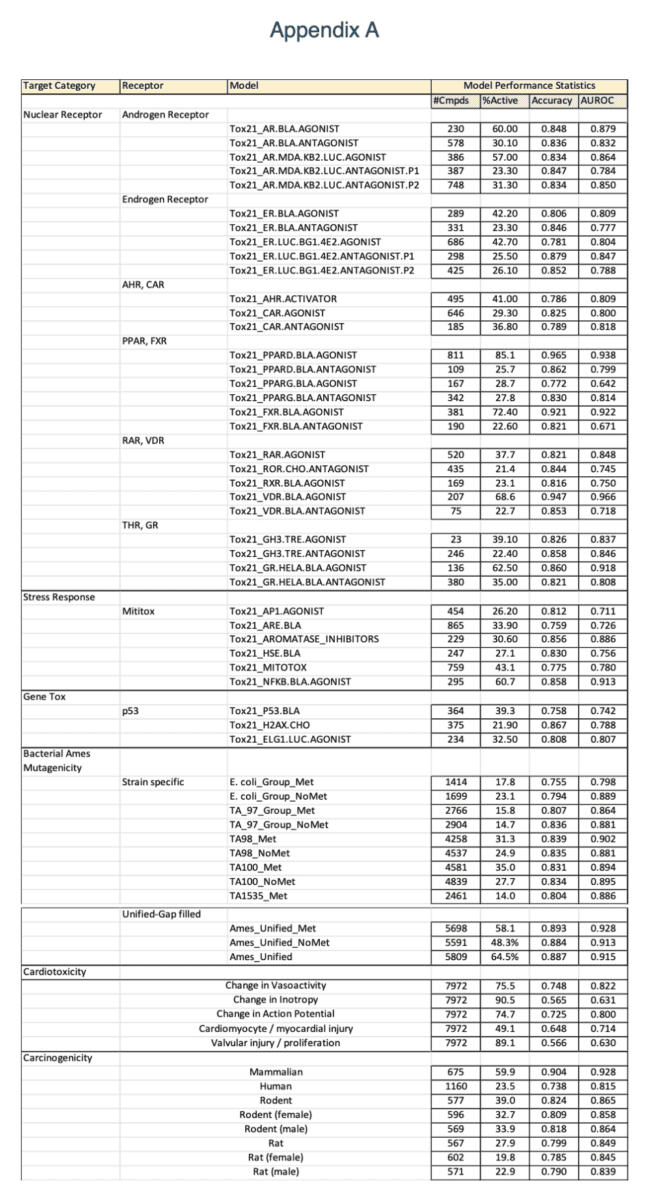
OrbiTox provides interpretable QSAR models for over 350 assays, including strain-specific Ames mutagenicity, human and rodent carcinogenicity with harmonized data calls, and predictions for cardiotoxicity failure modes from ToxCast and Tox21 assays. Each model has been systematically validated—with detailed methodologies described in peer-reviewed literature—and is built using state-of-the-art machine learning and AI techniques. Our models employ the latest Saagar descriptors to capture chemistry-aware functional groups and moieties, providing clear, chemistry-based reasoning for each prediction. These descriptors were rigorously compiled from open-source literature that correlates substructural features with physicochemical properties, ADME profiles, and toxicological endpoints. Moreover, OrbiTox visually highlights the key substructures influencing each prediction, empowering users to design new chemicals and generate mechanistic hypotheses. A listing and performance statistics for the in silico models currently available in the latest release of OrbiTox are shown in Appendix A.
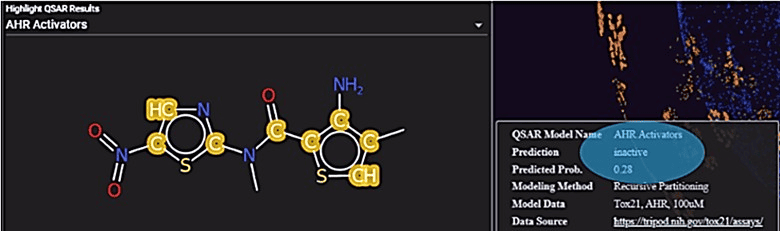
Multi-Domain Data Organization and Connectivity
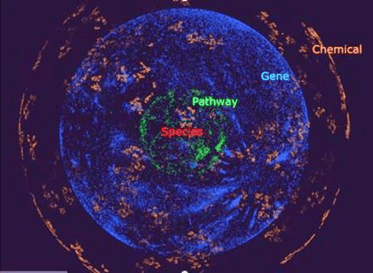
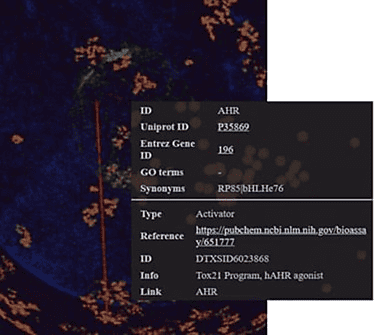
The OrbiTox database organizes its content into four clear layers—chemicals, genes, pathways, and species. For instance, it currently includes over 850K chemicals, 40,000 genes, 2,000 pathways, and 200 test organisms. The platform visually connects these layers using links based on both experimental and predicted data from QSAR models developed from trusted public data sources like Tox21, ToxNet, NTP reports, CPDB, IARC, and ToxCast assays. For example, an AhR activation test result links a chemical (orange layer) with the AhR target (blue layer), while a metabolic pathway connects related genes. This design offers fast, interactive 3D visualization and robust search capabilities, enabling users to easily explore complex relationships. OrbiTox also can securely integrate proprietary data with public datasets to support comprehensive translational discovery. It effortlessly handles large volumes of data while providing intuitive visualizations, enabling organizations to explore relationships between their own data and public information in a secure environment without compromising confidentiality.
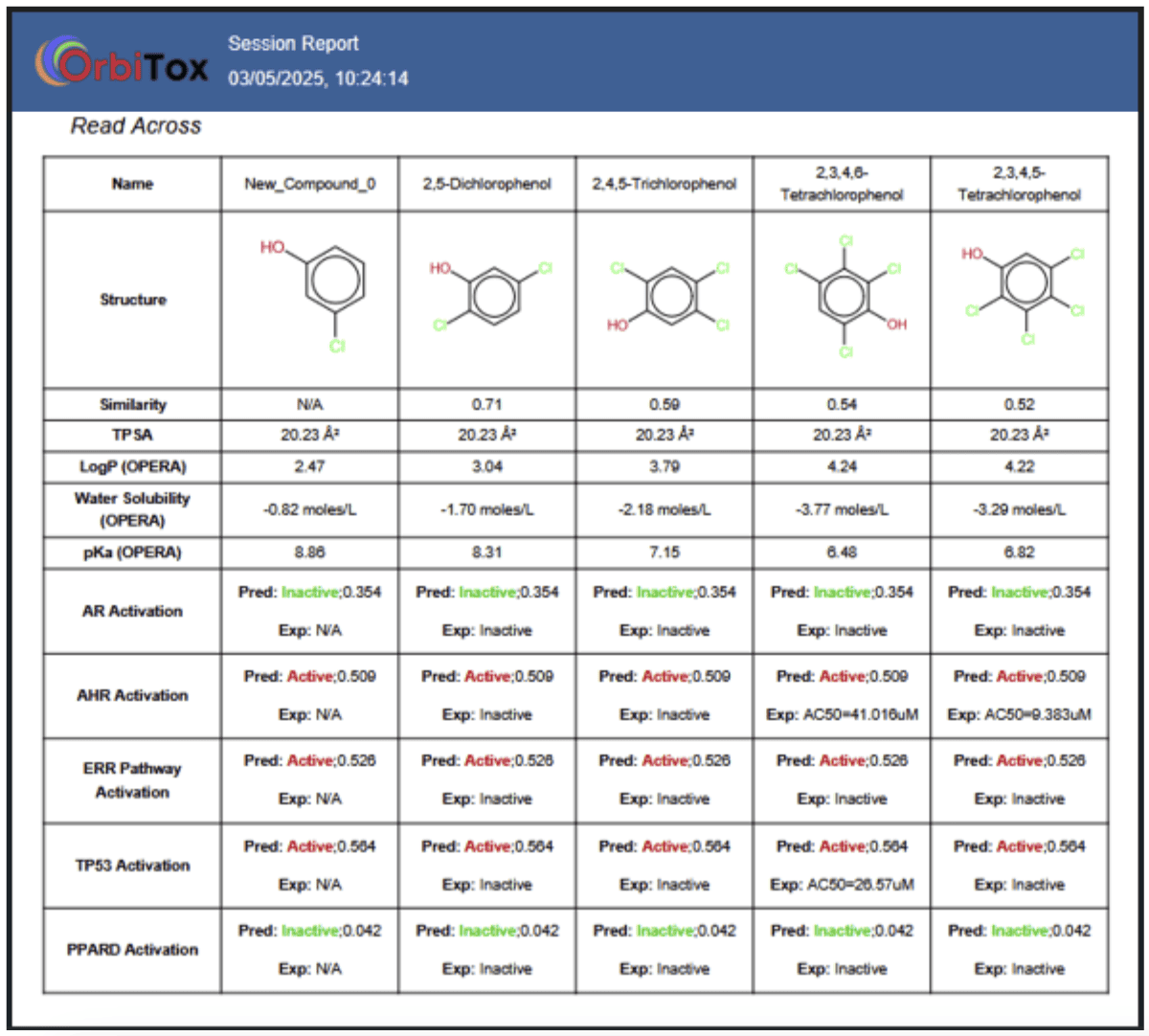
Data-Rich Read Across
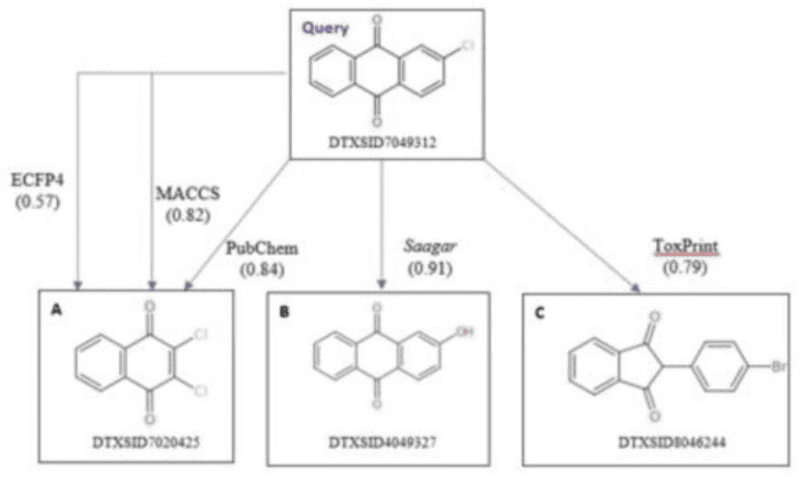 Read-across analysis reduces the need for additional experimentation by inferring data for a target molecule based on structurally similar source compounds. OrbiTox leverages Saagar descriptors to identify compounds with matching profiles across structure, physicochemical properties, bioactivity, and metabolism. Studies have shown that Saagar descriptors outperform traditional fingerprints like MACCS, ECFP4, PubChem, and ToxPrint—delivering improved scaffold similarity and accurate placement of key substituents. For instance, OrbiTox can instantly compare a query compound, such as Chloroanthraquinone, with AhR-active compounds from over 1,200 chemicals assayed in the Tox21 AhR assay using a modified Tanimoto index.
Read-across analysis reduces the need for additional experimentation by inferring data for a target molecule based on structurally similar source compounds. OrbiTox leverages Saagar descriptors to identify compounds with matching profiles across structure, physicochemical properties, bioactivity, and metabolism. Studies have shown that Saagar descriptors outperform traditional fingerprints like MACCS, ECFP4, PubChem, and ToxPrint—delivering improved scaffold similarity and accurate placement of key substituents. For instance, OrbiTox can instantly compare a query compound, such as Chloroanthraquinone, with AhR-active compounds from over 1,200 chemicals assayed in the Tox21 AhR assay using a modified Tanimoto index.
Bioactivity profiles for both target and source molecules are generated from a diverse set of QSAR models on OrbiTox (covering AR, AhR, ERR Pathway, TP53, and PPARD activation). For physicochemical and metabolic profiles, the platform integrates the open-source tools OPERA and SyGMa. Together, these features make OrbiTox a powerful and unique tool for selecting analogues in read-across assessments.
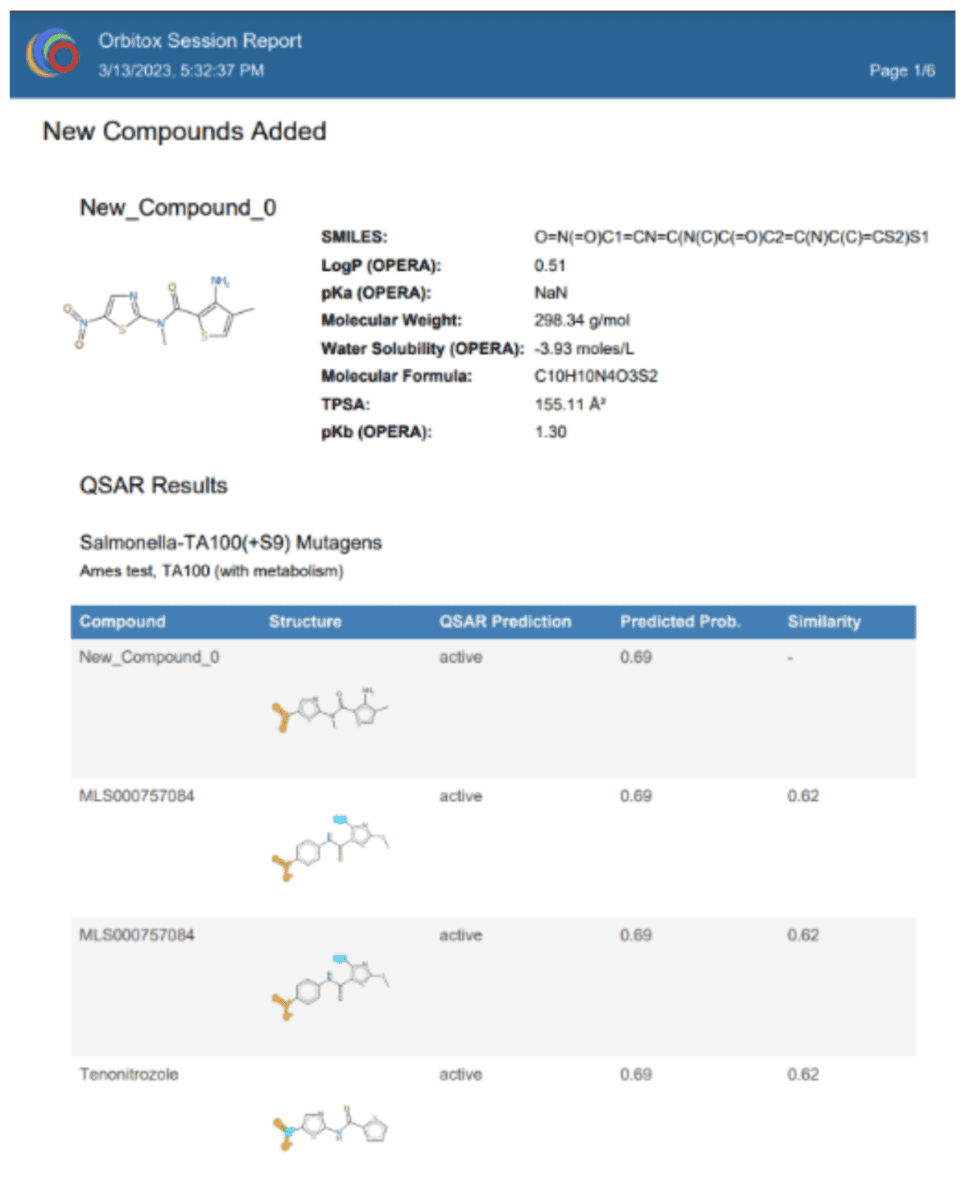
Advanced Reporting Features
OrbiTox features a robust reporting system that compiles all your interactive explorations and analyses into customizable reports. These reports can be saved in both tab-delimited format—for further analysis in other applications—and as PDF documents to serve as official records of your completed work. For instance, a sample PDF report displaying a predicted property profile shows computed molecular properties of the query structure, similar compounds with their Tanimoto similarity scores, and the predicted probability of a positive Ames test (using the Salmonella TA100 strain with metabolic activation). Another valuable report summarizes the read-across analysis by gathering the physicochemical, bioactivity, and metabolic profiles of structurally similar source chemicals. Additionally, to ensure full transparency and reproducibility, OrbiTox allows you to log the entire sequence of keystrokes used during your analysis.
Client-specific Customization
OrbiTox provides comprehensive, expert-curated data across various scientific domains relevant to toxicologists, along with advanced predictive tools such as instantaneous read-across and QSAR modeling. While the tool is highly effective right out of the box, we also offer several value-added services:
Incorporating Proprietary Data: Our flexible architecture allows you to integrate your organization’s proprietary chemical structures and property data into OrbiTox, establishing cross-domain connectivity.
Integrating Public Data: Our team expertly searches, cleans, and curates publicly available data to complement your proprietary datasets and expand the chemistry and property space.
Expanding Saagar Descriptors: Saagar descriptors, developed independently of third-party packages, can be extended using a semi-automated, data-driven approach to better cover your specific chemical space.
Enhancing QSAR Models: Once proprietary data is added, we can update existing QSAR models using a range of standard and cutting-edge modeling techniques. These models are rigorously benchmarked, validated, and deployed within your secure environment.
Building Custom QSAR Models: Our custom modeling pipeline combines automated and manual chemical curation with bespoke model development using advanced techniques, ensuring that the resulting models are robust, validated, and actionable.
Customizing Automated Reports: OrbiTox features a rich and flexible reporting system that you can tailor to your needs—whether you require detailed structural similarity reports, consensus predictions, visualizations of chemistry-based reasoning, or summaries of experimental and predicted data.
Here is a brief introductory video of OrbiTox
Publications and Presentations
- Ross, A., Gombar, V. & Sedykh, A. (2024). “OrbiTox – integration platform for chemical toxicity data. An update.” Poster presentation at the 63rd Annual Meeting of the Society of Toxicology, March 10-14, 2024, Salt Lake City, UT.
- Shah, R., Sedykh, A., Ross, A., Gombar, V. & Casey, W. (2023). “OrbiTox: An interactive predictive toxicology and translational discovery tool.” Oral presentation at the 12th World Congress on Alternatives and Animal Use in the Life Sciences, August 27-31, Niagara Falls, Canada.
- Gombar, V., Sedykh, A., Ross, A., Shah, R. & Casey, W. (2023). “OrbiTox: An interactive predictive toxicology and translational discovery tool.” Oral presentation at the 20th International Workshop on (Q)SAR in Environmental and Health Sciences, June 5-9, Copenhagen, Denmark.
- Gombar, V., Sedykh, A., Ross, A. Green, A., Shah, R. & Casey, W. (2023). “OrbiTox: Interactive Visualization of Multidomain Experimental and Predicted Data for Translational Discovery.” Oral presentation at the 62nd Annual Meeting of the Society of Toxicology, March 19-23, Nashville, TN.
- Sedykh, A., Ross, A., Gombar, V., Kleinstreuer, N., Merrick, A., Shah, R. & Casey, W. (2022). “OrbiTox – a computational translational discovery platform for data mining and read-across.” Oral presentation for the European Society of Toxicology In Vitro (ESTIV) 21st International Congress, November 21-25, Sitges, Spain.
- Ross, A., Sedykh, A.Y., Gombar, V.K., Kleinstreuer, N., Merrick, A., Shah, R.R. & Casey, W. (2022). “OrbiTox – interactive visualization of multi-domain experimental and predicted data for translational discovery.” Oral presentation for the 23rd European Society for Alternatives to Animal Testing (EUSAAT), September 26-28, Linz, Austria.
- Ross, A., Sedykh, A., Gombar, V., Kleinstreuer, N., Merrick, A., Shah, R. & Casey, W. (2022). “OrbiTox – A computational translational discovery platform for data mining and read-across.” Poster presentation at the 61st Annual Meeting of the Society of Toxicology, March 27-31, San Diego, CA.
- Gombar, V., Sedykh, A., Ross, A., Witt, K., Shah, R. & Casey, W. (2021). “OrbiTox – a translational discovery platform for concerted view and analysis of multi-domain data.” Poster presentation at the 10th Annual Meeting of the American Society for Cellular and Computational Toxicology (ASCCT), October 12-14, (Virtual).
- Sedykh, A., Ross, A., Shah, R., Witt, K., Casey, W. & Gombar, V. (2021). “OrbiTox – a platform for translational discovery and cheminformatics applications.” Poster presentation at the 19th International Workshop on (Q)SAR in Environmental and Health Sciences, June 7-9, (Virtual).
- Gombar, V., Sedykh, A., Ross, A., Witt, K., Shah, R. & Casey, W. (2021). “OrbiTox – A translational discovery platform for concerted view and analysis of big data.” Poster presentation at the 60th Annual Meeting of the Society of Toxicology, March 12-26, (Virtual).
- [2] Sedykh, A.Y.., Shah, R.R., Kleinstreuer, N.C., Auerbach, S.S. & Gombar, V.K. (2021). “Saagar-A New, Extensible Set of Molecular Substructures for QSAR/QSPR and Read-Across Predictions.” Chemical Research in Toxicology 34(2):634-640.
- [1] Sedykh, A., Shah, R., Kleinstreuer, N., Auerbach, S. & Gombar, V. (2020). “Interpretable QSAR and read-across predictions using Saagar – A new, extensible set of molecular substructures.” Poster presentation at the 19th International Workshop on (Q)SAR in Environmental and Health Sciences, Durham, NC.
