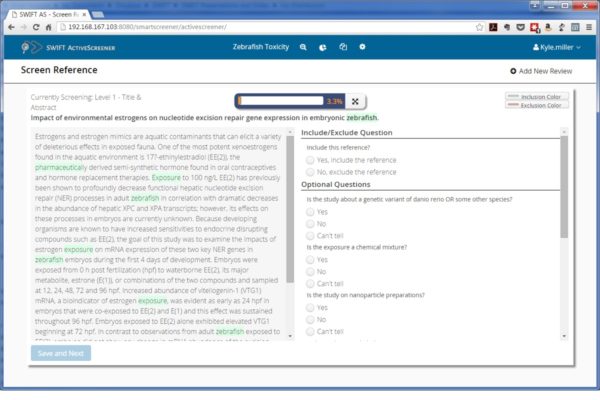
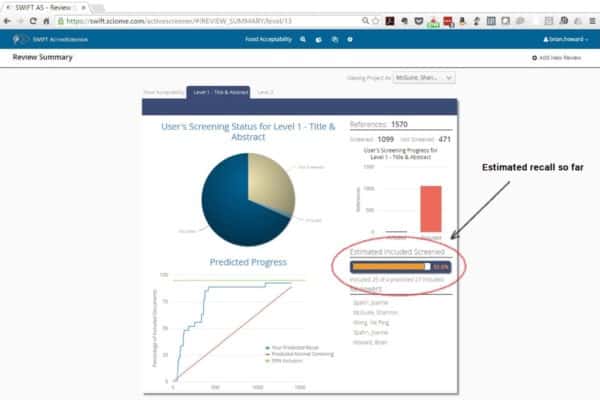
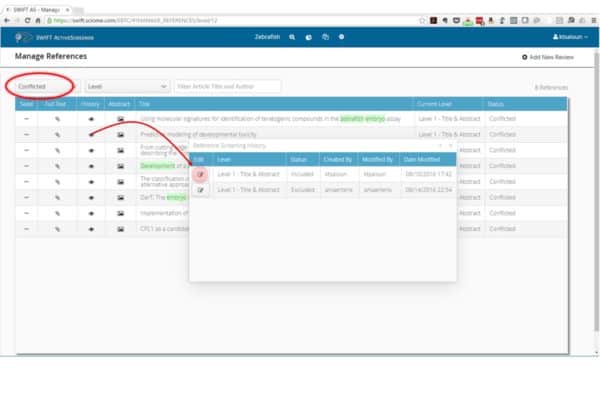
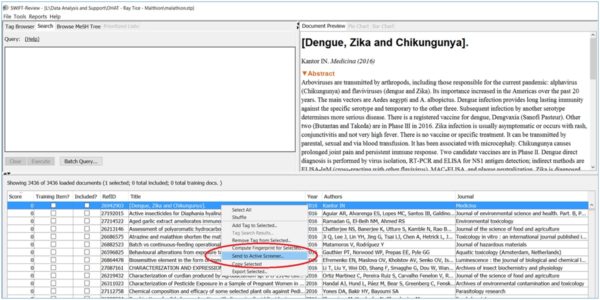
SWIFT-Active Screener is a web-based, collaborative systematic review software application. Active Screener was designed to be easy-to-use, incorporating a simple, but powerful, graphical user interface with rich project status updates. What makes Active Screener special, however, is its behind-the-scenes application of state-of-the-art statistical models designed to save screeners time and effort by automatically prioritizing articles as they are reviewed, using user feedback to push the most relevant articles to the top of the list.
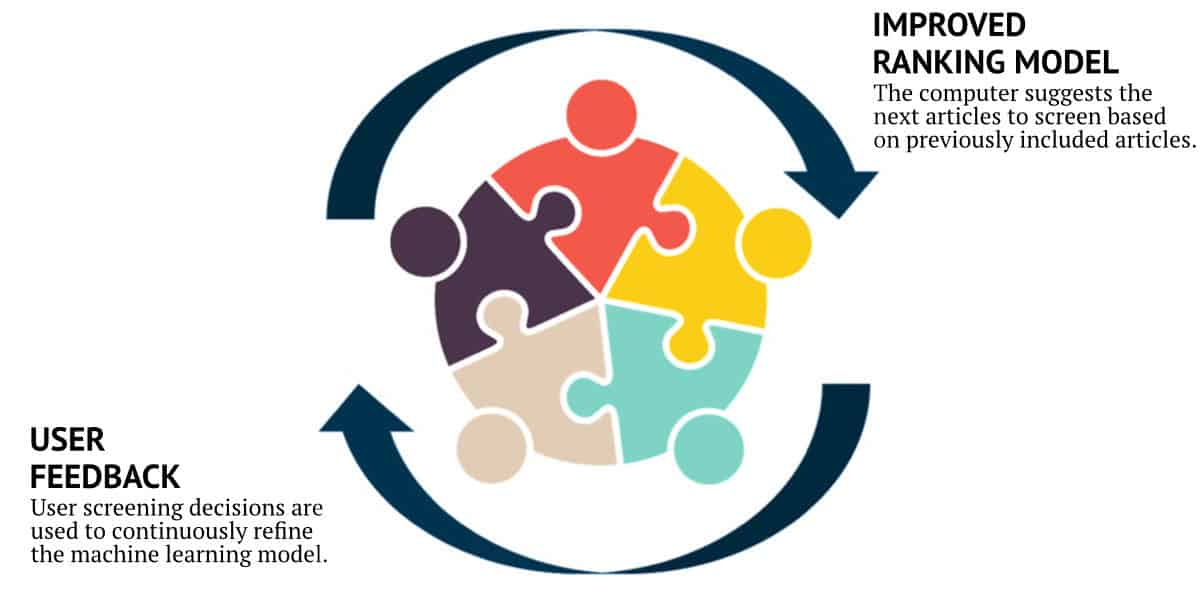
How Active Screener Works for You
As screening proceeds, reviewers include or exclude articles while an underlying statistical model automatically computes which of the remaining unscreened documents are most likely to be relevant. This “Active Learning” model is continuously updated during screening, improving its performance with each article reviewed. Meanwhile, a separate statistical model estimates the number of relevant articles remaining in the unscreened document list. Together, the combination of the two models allows users to screen relevant documents sooner and provides them with accurate feedback about their progress. Using this approach, the vast majority of relevant articles can often be discovered after reviewing only a fraction of the total number of articles. This can result in a significant time and cost savings for you and your team, especially for large projects.
For more information about SWIFT-Active Screener and other Sciome products and services, please contact us at swift-activescreener@sciome.com.
Data Integration
Active Screener integrates with all of the systematic review tools that you already use. It accepts imports from many of the most popular bibliographic databases and reference curation platforms including EndNote, Mendeley, Zotaro, PubMed, and SWIFT-Review. Results from screening activities in Active Screener can also be exported in standard data formats compatible with a wide range of applications including EndNote, Mendeley, Zotaro, PubMed, HAWC and Excel.
Publications, Presentations, Patents
- Wang A, Chappell GA, Varghese A, Howard BE, Schmidt L, Silva R, Arroyave W, Lunn, R, Rooney AA. (2024). “An Evaluation of the Performance of Survey Sampling in Systematic Evidence Mapping of Cancer Mechanistic Evidence: Polycyclic aromatic hydrocarbons (PAHs) and Key Characteristics of Carcinogens (KCCs) as a Case Study.” Poster presentation at the Society of Toxicology 63rd Annual Meeting and ToxExpo, Salt Lake City, UT.
- Howard BE, Norman CR, Tandon A, Shah R, Olker J, Hoff D (2023). “After Correcting for “Concept Drift,” Deep-Learning Methods Can Now Achieve Human-Level Performance When Predicting Article Exclusion Reasons During ECOTOXicology Knowledgebase Curation.” Poster presentation at the Society of Toxicology 62nd Annual Meeting and ToxExpo, Nashville, TN.
- Howard BE, Arroyave W, Schmidt L, Elmore B, Bisson W, Atwood S, Sethi M, Ewens A, Lunn R, Shah R, Merrick B.A., Wang A. (2023). “Use of Survey Sampling to Develop an Evidence Map of Carcinogenic Mechanistic Information for Polycyclic Aromatic Hydrocarbons (PAHs) and Key Characteristics of Carcinogens (KCCs).” Poster presentation at the Society of Toxicology 62nd Annual Meeting and ToxExpo, Nashville, TN.
- Howard BE, Norman CR, Tandon A, Shah R, Olker J, Hoff D (2022). “Improving the Efficiency of Literature Identification for the ECOTOXicology Knowledgebase Using Deep Learning.” Poster presentation at the Society of Toxicology 61st Annual Meeting and ToxExpo, San Diego, CA.
- Elmore R, Schmidt L, Lam J, Howard B, Tandon A, Norman C, Phillips J, Shah M, Patel S, Albert T, Taxman D, Shah R (2021). “Risk and Protective Factors in the COVID-19 Pandemic: A rapid Evidence Map (rEM).” Poster presentation at the Society of Toxicology 60th Annual Meeting and ToxExpo, Virtual.
- Howard BE, Norman CR, Tandon A, Shah R, Olker J, Elonen C, Hoff D (2021). “A Web-based Literature Identification Platform for the ECOTOXicology Knowledgebase, Powered by Deep Learning.” Poster presentation at the Society of Toxicology 60th Annual Meeting and ToxExpo, Virtual.
- Howard BE, Norman C, Tandon A, Shah R, Olker J, Elonen C, Hoff D (2020). “A Web-based Literature Identification Platform for the ECOTOXicology Knowledgebase, Powered by Deep Learning.” Platform presentation given at the SETAC North America 41st Annual Meeting, Virtual.
- Elmore R, Schmidt L, Lam J, Howard BE, Tandon A, Norman C, Phillips J, Shah M, Patel S, Albert T, Taxman DJ, Shah RR; (2020). “Risk and Protective Factors in the COVID-19 Pandemic: A Rapid Evidence Map.” Frontiers in Public Health 8 (787). doi: 10.3389/fpub.2020.582205.
- Howard BE, Phillips J, Tandon A, Maharana A, Elmore R, Mav D, Sedykh A, Thayer K, Merrick A, Walker V, Rooney A, Shah RR (2020). “SWIFT-Active Screener: accelerated document screening through Active Learning and integrated recall estimation.” Environment International 138 May 2020, 105623. doi: 10.1016/j.envint.2020.105623.
- Lam J, Howard BE, Thayer K, Shah RR (2020). “Evidence Mapping Health Outcomes: Rapid vs. Traditional Approaches.” Poster presentation at the Society of Toxicology 59th Annual Meeting and ToxExpo, Virtual.
- Howard B, Tandon A, Maharana A, Shah R (2019). “Using SWIFT-Active Screener to reduce the expense of evidence-based toxicology.” Presentation at the Cochrane Colloquium Santiago, Virtual.
- Lam J, Howard B, Bautz D, Anderson L, Tandon A, Maharana A, Shah R (2019). “Rapid Evidence Mapping for Environmental Toxicology.” Poster presentation at the 8th Annual Meeting of the American Society for Cellular and Computational Toxicology (ASCCT), Gaithersburg, MD.
- Lam J, Howard B, Thayer K, Shah R (2019). Low-Calorie Sweeteners and Health Outcomes: A Demonstration of rapid Evidence Mapping (rEM). Environment International 2019 Feb; 123:451-458. doi: 10.1016/j.envint.2018.11.070.
- Lam J, Howard BE, Thayer K, Shah RR (2019). “Rapid Evidence Mapping (rEM) in Environmental Health: Low-Calorie Sweeteners and Human Health. Poster presented at the Society of Toxicology 58th Annual Meeting and ToxExpo, Baltimore, MD.
- Lam J, Howard B, Tandon A, Maharana A, Elmore R, Merrick B, Shah R, Wolfe M (2019). “Rapid Evidence Mapping (rEM): Feminine Care Product Case Study.” Poster presented at the Society of Toxicology 58th Annual Meeting and ToxExpo, Baltimore, MD.
- Howard BE, Shah R, Mav D, Miller K (2019). “Methods and systems to estimate recall while screening an ordered list of bibliographic references.” Patent – US20190080002A1.
- Howard BE, Tandon A, Phillips J, Maharana A, Shah RR (2018). “Research Update: Using SWIFT-Active Screener to Reduce the Expense of Evidence Based Toxicology.” Poster presentation at the Fall meeting of the North Carolina Genetics and Environmental Mutagenesis Society (GEMS), RTP, NC.
- Lam J, Howard BE, Thayer K, Shah RR (2018). “Rapid Evidence Mapping for Health: A Case Study. Poster presentation at the Fall meeting of the North Carolina Genetics and Environmental Mutagenesis Society (GEMS), RTP, NC.
- Lam J, Howard B, Tandon A, Maharana A, Elmore R, Merrick B, Shah R, Wolfe M (2018). “Rapid Evidence Mapping (rEM): Feminine Care Product Case Study.” Poster presentation at the Fall meeting of the North Carolina Genetics and Environmental Mutagenesis Society (GEMS), RTP, NC.
- Howard BE, Tandon A, Phillips J, Shah M, Mav D, Shah R (2018). “Using Machine-Learning and SWIFT-Active Screener to Reduce the Expense of Evidence-Based Toxicology.” Poster presented at the Society of Toxicology 57th Annual Meeting and ToxExpo, San Antonio, TX.
- Lam J, Howard BE, Tandon A, Maharana A, Shah R (2018). “Rapid Evidence Mapping for Environmental Health.” Poster presented at the Society of Toxicology 57th Annual Meeting and ToxExpo, San Antonio, TX.
- Glynn C, Tokdar ST, Howard B, Banks, D (2018). “Bayesian Analysis of Dynamic Linear Topic Models.” Bayesian Analysis.
- Howard BE, Tandon A, Phillips J, Shah MR, Mav D, Shah RR (2017). “SWIFT-Active Screener: Faster Document Screening with Data Analytics.” Poster presentation at the Fall meeting of the North Carolina Genetics and Environmental Mutagenesis Society (GEMS), RTP, NC.
- Howard BE, Tandon A, Phillips J, Shah MR, Mav D, Shah RR (2017). “Using Machine Learning and SWIFT-Active Screener to Reduce the Expense of Evidence Based Toxicology.” Poster presentation at the Genetics and Environmental Mutagenesis 2017 Annual Fall Meeting, RTP, NC
- Howard BE, Miller K, Phillips J, Shah M, Mav D, Thayer K, Shah R (2017). “Enabling Evidence-Based Toxicology with SWIFT Active Screener.” Poster presentation at the Society of Toxicology’s 56th Annual Meeting and ToxExpo, Baltimore, MD.
- Miller K, Howard BE, Phillips J, Shah MR, Mav D, Thayer K, Shah RR (2016). “SWIFT-Active Screener: Reducing Literature Screening Effort Through Machine Learning for Systematic Reviews.” 24th Cochrane Colloquium. Seoul, South Korea.
- Howard BE, Miller K, Shah RR (2016). “SWIFT-Active Screener (A Brief Introduction).” National Institute of Environmental Health Sciences (NIEHS). RTP, NC.
- Miller K, Howard BE, Phillips J, Shah MR, Mav D, Thayer K, Shah RR (2016). “Updating Systematic Reviews with Active Learning.” 24th Cochrane Colloquium. Seoul, South Korea.
- Miller K, Howard BE, Phillips J, Shah MR, Mav D, Shah RR (2016). “SWIFT-Active Screener: Reducing Literature Screening Effort Through Machine Learning for Systematic Reviews.” Poster presentation at the Society of Toxicology’s 55th Annual Meeting and ToxExpo, New Orleans, LA.
Current Users Include





Another important aspect is that the SWIFT team is very committed to the development of the application and to the users. I strongly recommend SWIFT Active Screener.