FIDDLE
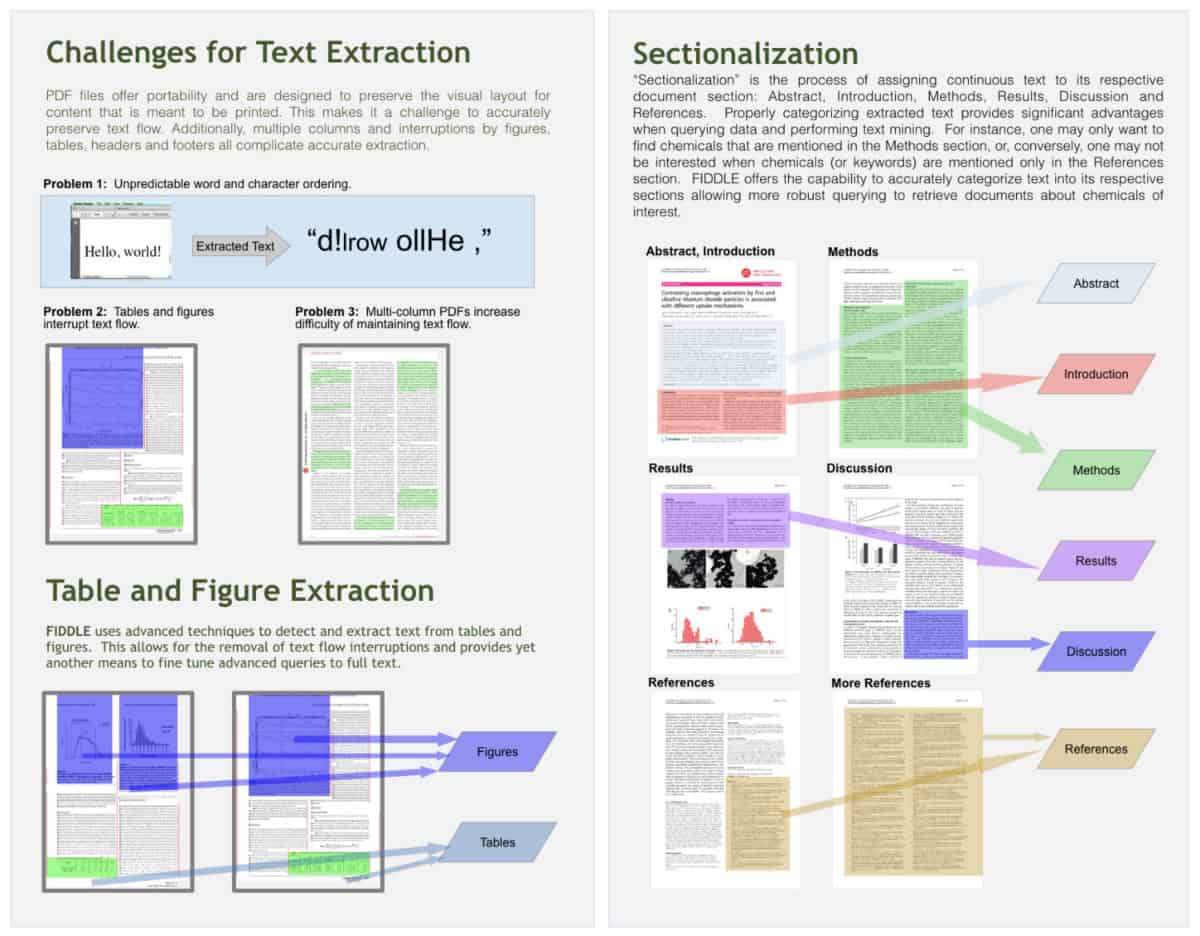
The ‘Flow-Intelligent Document Decoder for Literature Extraction’ (FIDDLE), is a novel algorithm and a tool under development, which can accurately extract text from scientific PDF documents. In addition to preserving the correct word order across columns, and even around figures and tables, FIDDLE is also able to accurately sectionalize scientific documents in an automated fashion. When key scientific terms (e.g. chemical names) are mentioned in different document sections (e.g. title, abstract, methods, references, etc.) they may carry greatly different implications. Therefore, the ability to correctly divide documents into meaningful sections is a critical capability in the context of literature mining.
When combined with a custom built, dictionary-based chemical name recognizer, FIDDLE is able to accurately extract chemical names from full text PubMed manuscripts at a rate several times greater than would otherwise be possible using only the associated MeSH terminology or the text of the titles and abstracts. Our approach offers the ability to perform full text extraction of scientifically relevant keywords (e.g. chemical name or phenotypic endpoint) and in turn greatly enriches text and literature mining capabilities with wide ranging applicability in the environmental and health sciences.
Publications, Presentations, Patents
- Howard, BE (2022). “Towards Automating Information Extraction with FIDDLE 2.0: From Text Annotation to Interoperable Information Extraction via Machine Learning.” National Academies of Sciences, Engineering and Medicine (NASEM) Workshop to Support EPA’s Development of Human Health Assessments: Artificial Intelligence and Open Data Practices in Chemical Hazard Assessment (Virtual).
- Howard BE, Tandon A, Norman C, Albert T, Patel S, Elmore R, Schmidt L, Shah R (2021). “Towards Automating Information Extraction with FIDDLE: From Text Annotation to Interoperable Information Extraction via Machine Learning.” Oral presentation at the 10th Annual ASCCT Meeting (Virtual).
- Howard BE (2021). “Towards Automatic Information Extraction with FIDDLE 2.0: From Text Annotation to Interoperable Information Extraction via Machine Learning.” Oral presentation at the OpenTox Virtual Conference (Virtual).
- Howard BE, Maharana A, Tandon A, Albert T, Phillips J, Taylor M, Thayer K, Shah R (2021). “Semi-automated Data Extraction Workbench for Environmental Health.” Poster presentation at the Society of Toxicology 60th Annual Meeting and ToxExpo (Virtual).
- Howard BE, Maharana A, Tandon A, Shah R (2019). “Semi-automated Data Extraction Workbench for Environmental Health.” Poster presentation at the Society of Toxicology 58th Annual Meeting and ToxExpo, Baltimore, MD.
- Phillips J, Shah R, Howard, B (2019). Methods and systems for efficient and accurate text extraction from unstructured documents. US Patent 10,360,294; 2019/07.
- Howard BE, Phillips J, Tandon A & Shah RR (2018). “PDF Text Extraction with FIDDLE.” Oral presentation at the National Toxicology Program (NTP), RTP, NC.
- Phillips J, Howard BE & Shah R (2014). “Scientific Text Extraction Using FIDDLE: A Foundation for Accurate Literature Mining.” Poster presentation at the US EPA ToxCast Data Summit, RTP, NC.
P-MACD
DMR Generator Tool
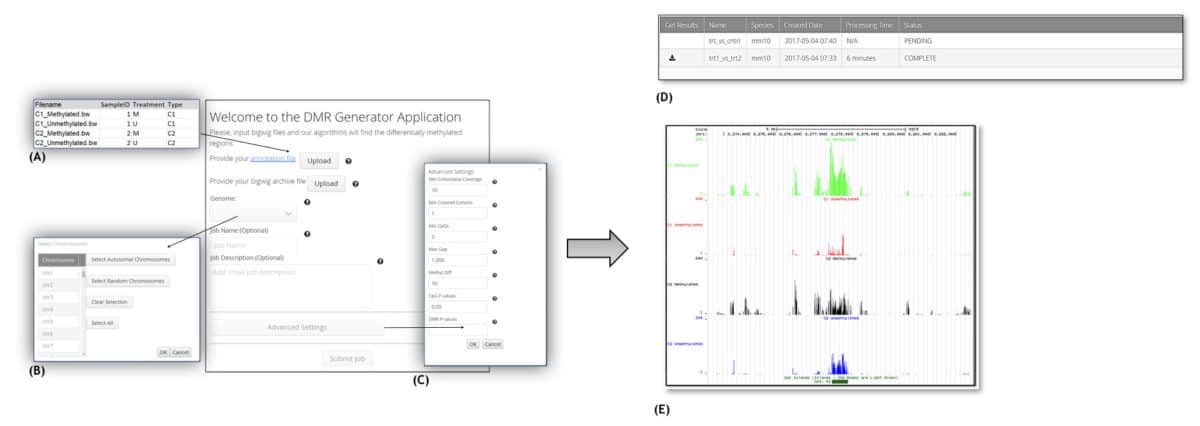
Recent improvements in next-generation sequencing technology allow for the efficient, genome-wide measurement of changes in DNA methylation status. With diverse applications including novel research into the mechanisms, regulation and biological consequences of genomic imprinting, chromosomal stability and embryonic development, these technologies have opened up new frontiers in a multiplicity of domains including toxicology, pharmacology, medicine and genetics. Unfortunately, however, the statistical methods available for analyzing the resulting data have so far been quite limited, and to date there are few existing methods that can accurately identify differentially methylated regions (DMR) from raw sequencing data. To overcome this limitation, Sciome has developed DMR Generator, a web based application for conducting a bioinformatics analysis of whole-genome DNA methylation profiles. In addition, DMR Generator also provides the opportunity for integrated downstream data analysis of the results.
DMR Generator application workflow: A) Upload bigWig data and annotation file; B) select genome and chromosomes; C) (optionally) specify advanced settings; D) monitor progress and download results. E) Resulting DMRs can be plotted in a genome browser.
Publications and Presentations
- Li R, Grimm SA, Mav D, Gu H, Djukovic D, Shah R, Merrick A, Raftery D and Wade PA, “Obesity Predisposes to Colon Cancer by Reprogramming Colonic Cell Metabolism and Rewiring Colonic Signal Transduction in Mice.” (Submitted to Gastroenterology).
- Grimm SA, Shimbo T, Mav D, Takaku M, Thomas JW, Auerbach S, Bennett BD, Bucher JR, Burkholder AB, Dai S, Du Y, French JF, Li J, Merrick AB, Tice RR, Wang T, Xu X, Shah R, Bushel PR, Fargo DC, Mullikin JC, Wade PA, “Transcription factor-DNA interactions govern allelic inheritance of DNA methylation in mice. (In preparation)
- Mav D, Tandon A, Phillips J, Howard BE, Wade P, Shah R (2017). DMR Generator: Interactive Application for Differentially Methylated Regions Detection. In: 2017 Annual Meeting Abstract Supplement, Society of Toxicology, 2017.
- Tandon A, Mav D, Phillips J, Shah M, Phadke D, Howard BE, Merrick BA, Wade P, Bucher B, Shah R. DMR Generator: Interactive Application for Differentially Methylated Regions Detection. (Submitted to Bioinformatics)
Risk21 RoadMap
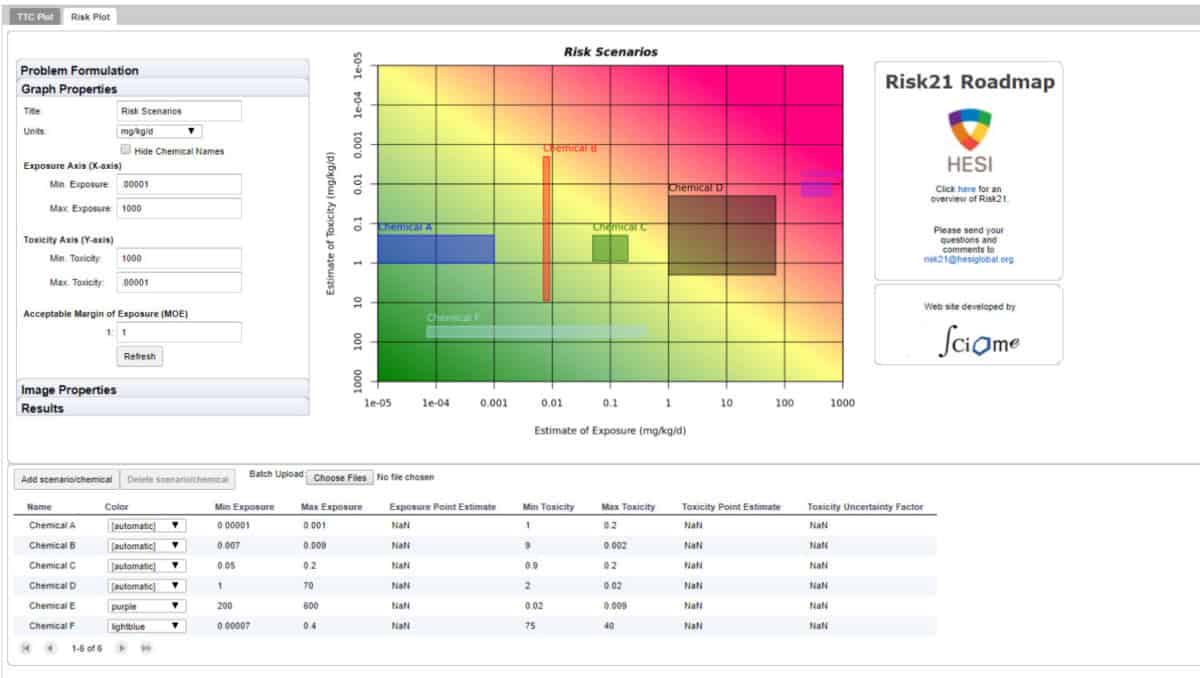
The Risk21 RoadMap is a web-enabled visualization tool developed at Sciome in collaboration with the ILSI Health and Environmental Sciences Institute (HESI). This tool automates the Risk21 framework for problem formulation and risk assessment, as developed by researchers at HESI. The Risk21 framework is a highly visual process for problem formulation and risk assessment that allows researchers to integrate available exposure and toxicology information in way that can be effectively communicated with a variety of stakeholders, including those without extensive training in the toxicological sciences. Sciome’s role in this project was to design and implement the software on behalf of the client. The resulting application, referred to as the Roadmap Risk21 Visualization (RRV) tool, is currently being utilized by hundreds of research groups across the globe. A number of stakeholder engagement and educational workshops within the US and around the world have also been conducted, focusing on the use of RRV for toxicological research.
Publications and Presentations
- Wolf DC, Bachman A, Barrett G, Bellin C, Goodman JI, Jensen E, Moretto A, McMullin T, Pastoor TP, Schoeny R, Slezak B, Wend K. Embry MR. (2016). Illustrative case using the RISK21 roadmap and matrix: prioritization for evaluation of chemicals found in drinking water. Critical Reviews in Toxicology, 46: 43-53.
- Embry MR, Bachman AN, Bell DR, Boobis AR, Cohen SM, Dellarco M, Dewhurst IC, Doerrer NG, Hines RN, Moretto A, et al. (2014). Risk assessment in the 21st century: Roadmap and matrix. Critical Reviews in Toxicology, 44:6-16.